Our assay platform demonstrates that receptor expression levels influence the pharmacological profiles of 5 HT2A/2B ligands, unmasking partial agonism and emphasising the need to account for receptor density and functional reserve to avoid underestimating partial agonism and potential off target toxicity in pre clinical screens.
The 5-HT2 receptor family comprises three closely related GPCRs (5-HT2A, 5-HT2B and 5-HT2C) which bind the endogenous neurotransmitter serotonin (5-Hydroxytryptamine or 5-HT). The 5-HT2A receptor is expressed abundantly in the CNS and is an attractive clinical target for anxiety, psychosis and OCD. However, selectivity is an important consideration when developing ligands for this target, as the related 5-HT2B receptor is expressed in the cardiovascular system and the chronic activation of this receptor by unselective or poorly selective 5-HT2A agonists is known to induce adverse cardiopulmonary effects.
There are numerous known ligands for the 5-HT2A receptor, however many of these lack specificity, exhibiting appreciable off-target activity at 5-HT2B.
It has been reported that differences in GPCR expression levels and the presence of receptor reserves can alter both the receptor coupling efficiency and the apparent pharmacological profile of 5-HT2A receptor ligands (1,2). It can therefore be challenging to fully characterise the pharmacology of ligands for this receptor, necessitating careful evaluation in orthogonal assays.
BioAscent scientists generated multiple HEK-293T cell lines stably expressing human 5-HT2A or 5-HT2B receptors. Receptor expression levels were quantified in all the clones by RT-qPCR, and three clones per receptor were selected, representing a range of expression levels, for full pharmacological characterisation using the FLIPR intracellular Ca2+ mobilisation assay and xCELLigence label-free cellular impedance assay (Figure 1).
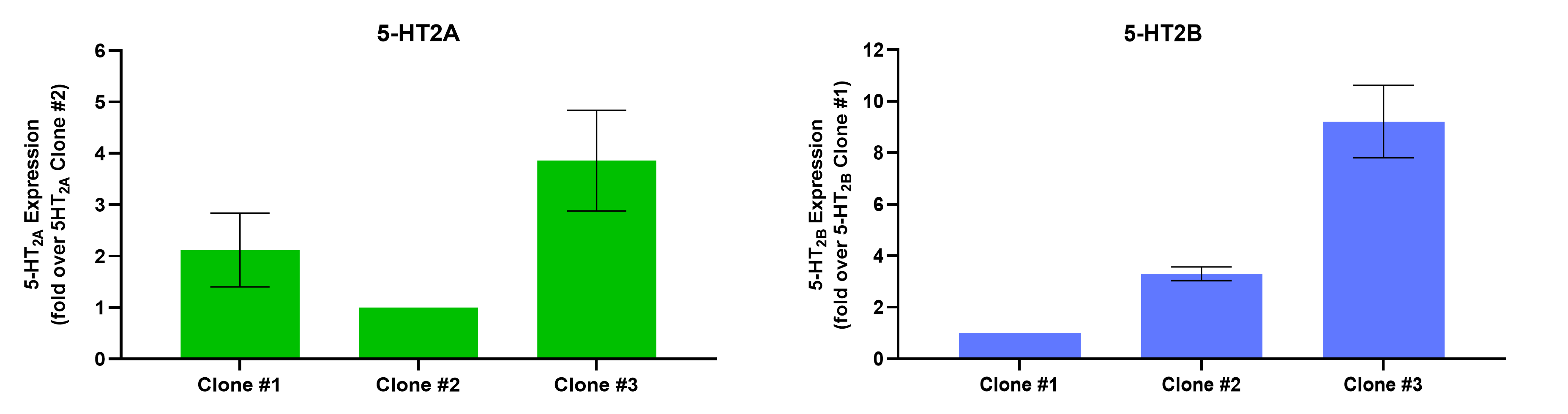
Figure 1. Quantification of 5-HT2A/5-HT2B receptor expression by RT-qPCR. 10ng of cDNA was used for qPCR with PowerUp™ SYBR Green Master Mix. Transcript levels were normalised to GAPDH and expressed relative to the lowest expressing clone using the 2-ΔΔCt method. Mean triplicate expression data ± SEM is shown. (n=3/N=3)
Expression-dependent potency and efficacy profiles at 5-HT2A and 5-HT2B receptors
The endogenous 5-HT2 agonist, 5-HT, and psilocin, a CNS-permeant compound with known partial agonist activity at 5-HT2A, were profiled in the FLIPR assay (Figure 2). 5-HT demonstrated full agonist activity across all three 5-HT2A receptor clones, although there were obvious differences in potency for each clone correlating to receptor expression levels. In contrast, psilocin displayed variable activity depending on receptor expression. Partial agonism was observed in clones #1 and #3 but the compound was inactive against clone #2. Our data highlights the importance of considering receptor expression levels when profiling ligands for 5-HT2A. Psilocin is a well-documented partial agonist; however, this activity could be completely missed if screening compounds solely in a lowly expressing clone. For those clones which did show activity, there were marked differences in observed Emax and potency.
Similarly, when screened against 5-HT2B, 5-HT exhibited potent agonism for all three clones, whereas psilocin displayed partial activity exclusively for clone #3 and was inactive against the clones #1 and #2.
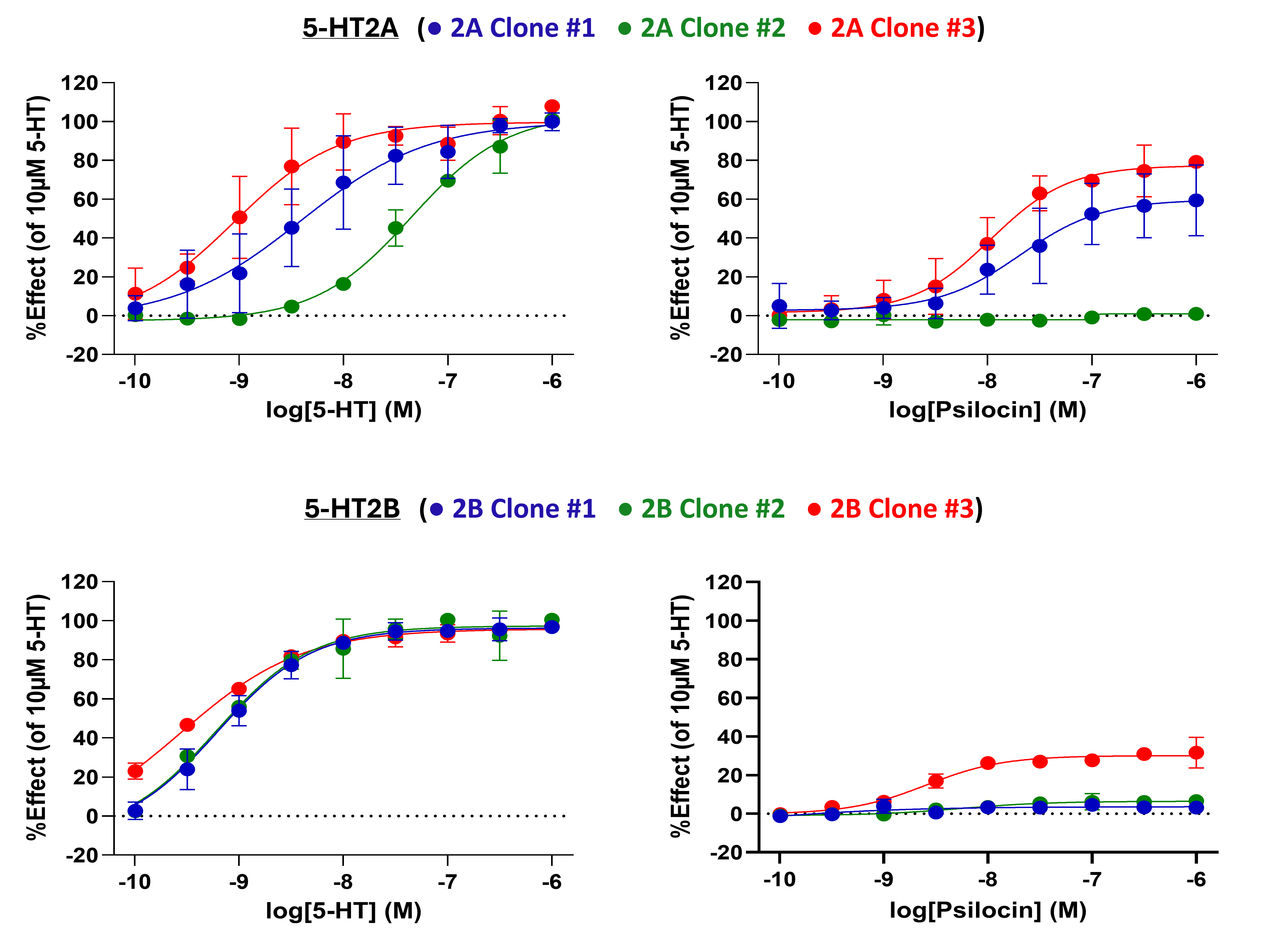
Figure 2. 5-HT2 receptor agonists show expression-dependent potency and efficacy profiles at 5-HT2A and 5-HT2B receptors. Agonist DRCs were normalised to DMSO (min control) and 10µM 5-HT (max control) and plotted for 5-HT2A and 5-HT2B stable cell clones. Mean duplicate data from three independent runs (n=2/N=3) is shown ± SEM.
Correlation between assay technologies
Correlation between technologies was studied by screening 7 known agonists with varying activity at 5-HT2A and 5-HT2B, including 5-HT and psilocin, in two assay formats. Compounds were screened in both the FLIPR assay and in the label-free xCELLigence cellular impedance assay, which was used as an orthogonal technology. Agonist potencies (pEC50) were found to be consistent between both assays, demonstrating inter-technique consistency for ligand characterisation (Figure 3).
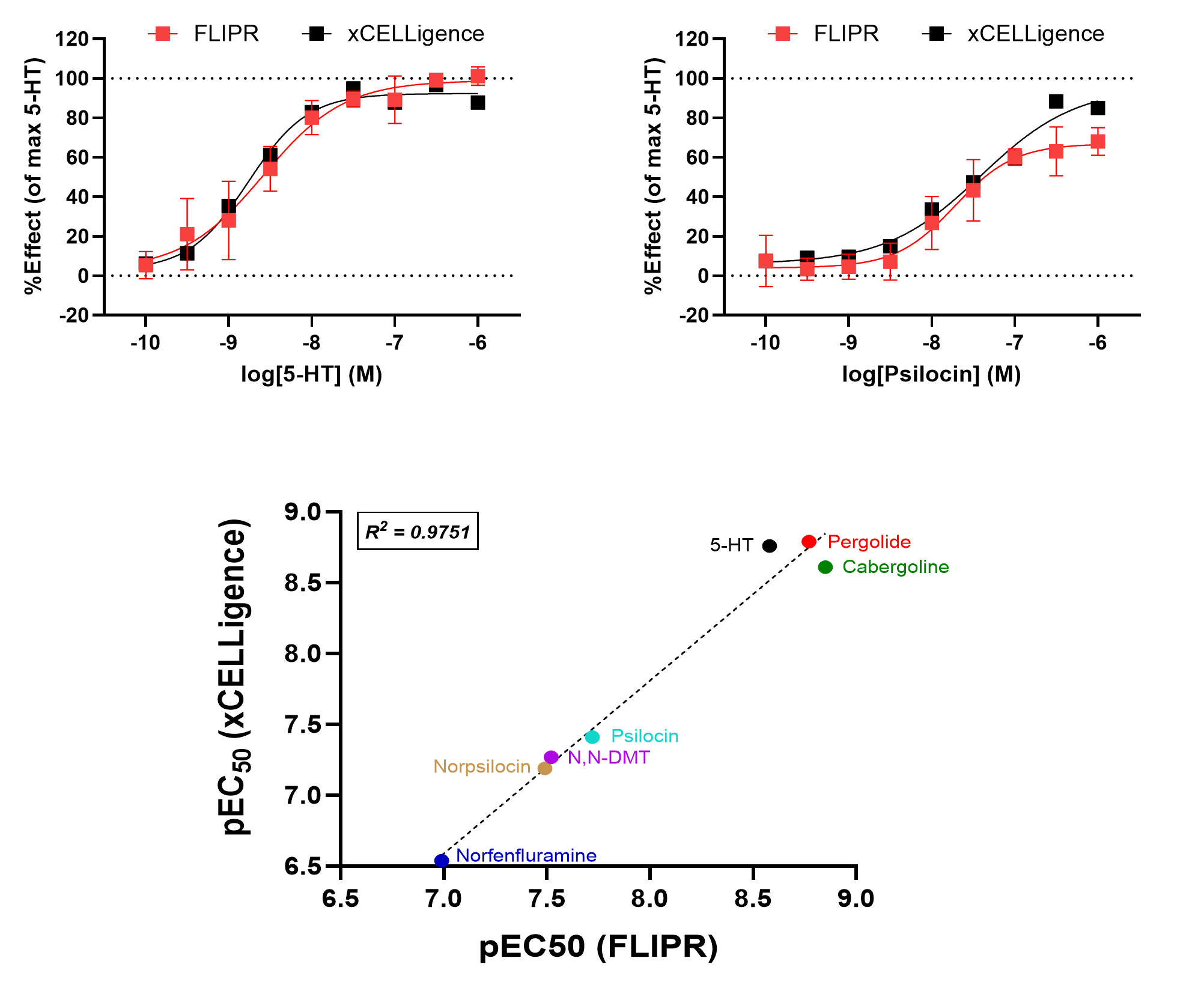
Figure 3. 5-HT2A agonists show similar activity across both FLIPR and xCELLigence technology assays. Agonist activity was normalised to DMSO (“0% effect”) and 10µM 5-HT (“100% effect”) and plotted as dose-response curves for both assays. Agonist potency (pEC50) values were calculated and correlated for all agonists between the two assays.
These data show that differences in receptor expression levels significantly influences the pharmacological profiles of 5-HT2A/2B ligands and unmasks partial agonist activity in ligands which have shown inconsistent activity in the literature. These results potentially explain the mixed literature reports and underscore the importance of carefully considering receptor density and functional reserve when profiling for selectivity. Failure to account for these factors leads to the danger of underestimating partial agonism and potentially off-target toxicity in pre-clinical screens.
These findings highlight the challenges in developing truly selective ligands for the 5-HT2A receptor. Furthermore, they underscore the importance of incorporating multiple physiologically relevant models and orthogonal assay approaches into pre-clinical drug safety and efficacy profiling, to ensure accurate predictions of clinical outcomes.
For full detail of all the methods and information on additional experiments, please see our posters on this work:
Learn more about BioAscent’s assay development and screening expertise here.
Jacobson, K.A. (2015) Biochem Pharmacol. https://doi.org/10.1016/j.bcp.2015.08.085
Li et al. (2023) PLoS One. https://doi.org/10.1371/journal.pone.0283477




